Autor de la sección: Sebastian Jentschke
How do I identify outliers and filter them out from being used in analyses?¶
- open the
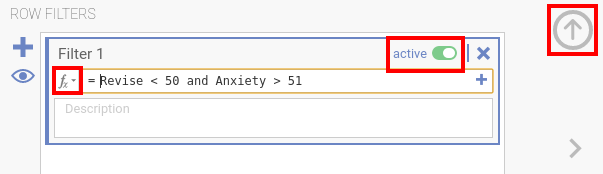
Datatab and selectFilter(either by using the symbol in the icon bar or the one in the bottom-left corner of the jamovi window)in order to access functions, press thefxicon in the filter settingsthere also is a switch where you can activate or deactivate the filter (see the comment in red below)you close the filter settings by pressing the arrow in the top-right corner - there are three large approaches, to exclude outliers:
- based upon z-scores (the absolute value should be larger 3.3; this equals to a probability of 0.1% = 1 / 1000; based upon a standard normal distribution ~ parametric)
- based upon the IQR (like in a box plot; based upon ranks and quantiles ~ non-parametric)
- based on the Mahalanobis distance (multivariate outliers)
for 1. and 2., there exist functions in jamovi (see next bullet points), for 3. you have to use R-code (decribed two bullet point below); for 2. you could also do it visually (three bullet points below) - you can either use an function-based selection; the functions below filter out lines based on either the z-scores (first line), the interquartile range (IQR, second line) or by excluding certain rows / row numbers (e.g., based upon the output from the calculation of the Mahalanobis distance further below; third line):
MAXABSZ([VARIABLE1], [VARIABLE2], …)
MAXABSIQR([VARIABLE1], [VARIABLE2], …)
IFMISS(MATCH(ROW(), [ROWNUMBER 1], [ROWNUMBER 2], …), 1, 0)
 the following code example detects multivariate outliers based upon the Mahalanobis distance (remember to adjust the variable names in VL)
the following code example detects multivariate outliers based upon the Mahalanobis distance (remember to adjust the variable names in VL)# this list should contain the names of your INDEPENDENT VARIABLES # you should not include your dependent variables # if you already use a filter set it to inactive # hint: you can get the names of your variable with names(data) # the syntax is adjusted for jamovi (the data frame is called data, # but can easily be used within R by just changing data to the name of your data frame VL = c('dan.sleep', 'baby.sleep', 'day') # brief explanation: the code calculates the Mahalanobis distance for all variables in VL, # then calculates the p-value (pchisq) and show lines with variables that had a p-value < 0.001 row.names(data)[ pchisq(unname( mahalanobis(data[, VL], colMeans(data[, VL]), cov(data[, VL]))), df=length(VL), lower.tail=FALSE) < 0.001]
the output from theR-code tells you which lines you should de-selectyou use the scripts within the Rj editor, just copy-and-paste them and run them by hitting the ►-button (the little green triangle) - the filter conditions can then be combined using boolean
and/or:MAXABSZ([VARIABLE1], [VARIABLE2], …) < 3.3 and MAXABSIQR([VARIABLE1], [VARIABLE2], …) < 3 and IFMISS(MATCH(ROW(), [ROWNUMBER 1], [ROWNUMBER 2], …), 1, 0)
- instead of using the second line (
MAXABSIQR) you could also de-select cases by excluding their respective row numbers in the dataset (as in the third line; you would then visually check the outliers in the box-plots underDescriptives, ensuring that the tick boxLabel outliersis set and exclude the row numbers that are marked as outliers)